Classification of viruses: Baltimore& ICTV

Naming viruses and placing them in a taxonomic system is called the classification of viruses. It is always controversial while defining and classifying viruses because they are non-living particles with some chemical characteristics similar to those of life or non-cellular life. They do not fit into the biological classification system established for cellular organisms due to the non-living characteristics they have.
Basis of classification of viruses
Viruses are classified mainly by the following characters.
- Their Phenotypic characteristics, such as morphology
- The nucleic acid type which they have i.e. single-stranded or double-stranded DNA or RNA
- Their mode of replication within the host
- Type of host organism i.e. bacteria, plant, animal cell, etc.
- Type of disease they cause in their host
But now, with the advancement in genomic sciences viruses are generally classified on the basis of their Nucleic acid type. Two well-known systems of classification of viruses are named right below.
- ICTV virus Classification 1st published in 1970 (International Committee on Virus Taxonomy)
- The Baltimore classification in 1971
The Baltimore classification
The Baltimore classification, developed by David Baltimore in 1971, is a virus classification system that divides viruses into families, depending on their
- Type of genome (DNA, RNA, single-stranded (ss), double-stranded (ds)
- Their method of replication
Viruses placed in a given category will all behave in much the same way, which can point out further the characters of the newly discovered viruses in a specific group.

Seven Baltimore classes of the viruses are given below.
I: Double stranded (ds) DNA viruses ( for example Adenoviruses, Herpesviruses, Poxviruses)
II: Single stranded (ss) DNA viruses (+ strand or “sense”) DNA (for example Parvoviruses)
III: Double stranded (ds) RNA viruses (for example Reoviruses)
IV: Single stranded RNA viruses with positive sense strand (+ssRNA) (for example Coronaviruses, Picornaviruses, Togaviruses)
V: Single stranded RNA viruses with negative sense strand (-ssRNA) RNA (for example Orthomyxoviruses, Rhabdoviruses)
VI: ssRNA-RT viruses (+ strand or sense) RNA with DNA intermediate in life-cycle (for example Retroviruses)
VII: dsDNA-RT viruses DNA with RNA intermediate in life-cycle (for example Hepadnaviruses)
Group I: Double-stranded DNA viruses
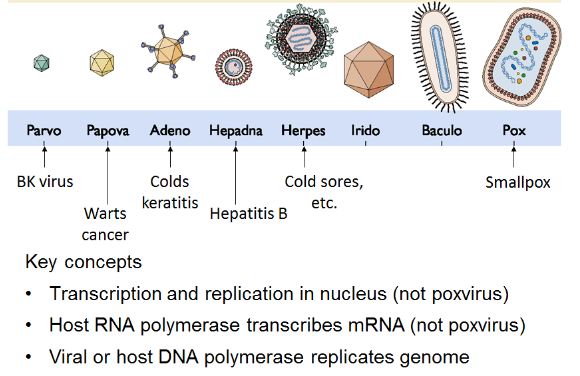
These types of viruses enter the host cell nucleus before they are able to replicate. Furthermore, these viruses need host cell polymerases enzymes for replication of the viral genome and, so, are heavily dependent on the host cell cycle. For accurate infection and replication of virus the host cell should be in dividing stage as during replication the cell’s polymerases enzymes are active. The virus may also forcibly induce the cell to undergo cell division, which may lead to the transformation of the normal cell and to cancerous cell mass. Examples include Herpesviridae, Adenoviridae, and Papovaviridae.
Poxvirus family that infects vertebrates and causes the smallpox disease is the only one example in which a class I virus is not replicating within the nucleus
Group II: Single-stranded DNA viruses
Group II includes the, Circoviridae, Anelloviridae and Parvoviridae (that infect vertebrates animals), the Microviridae (infect prokaryotes) and the Nanoviridae and Geminiviridae (that infect plants). Majority of these have circular genomes.
Eukaryote-infecting viruses replicate mostly within the nucleus by a rolling circle mechanism. By this all viruses in this group form a “double stranded DNA intermediate molecule” during their genome replication. This is normally created from the viral DNA with the help of the host’s own DNA polymerase.
In 1959, Sinshemer working with phage Phi X 174 showed that they possess single-stranded DNA genomes. Despite this discovery, until relatively recently it was believed that most DNA viruses contained double-stranded DNA.
But today it is well discovered that single-stranded DNA viruses can be highly abundant in the aquatic ecosystems, sediments, terrestrial environments, as well as metazoan-associated and marine microbial mats. Many of these “environmental” viruses belong to the family Microviridae.
Group III: Double-stranded RNA viruses
Double-stranded (ds) RNA viruses are a diverse group of viruses that infect a large range of hosts like animals, bacteria, plants and fungi. Members of this group include the rotaviruses, globally known as a common cause of gastroenteritis in kids, and bluetongue virus, an economically damaging pathogen of cattle and sheep.
This group includes number of families in which two major families, the Reoviridae and Birnaviridae are well known. Of these families, the Reoviridae is the largest and most diverse in terms of host range.
- These viruses are all nonenveloped and have icosahedral capsids and segmented genomes.
- Like the most RNA viruses, these viruses replicates in the “Core” capsid present in the cytoplasm of the host and do not use the replication polymerases of the host cell.
- In these, replication is monocistronic and includes individual segmented genome, which means that each of the genes codes for only one protein, unlike other viruses that exhibit more complex translation.
Group IV & V: Single-stranded RNA viruses
These single stranded RNA viruses belong to Class IV or V of the Baltimore classification. They could be grouped into positive sense or negative sense according to the sense or polarity of RNA molecule. The single stranded RNA is the common feature of these viruses. The replication of viruses happens in the cytoplasm or nucleus of the host cell. Class IV and V ssRNA viruses do not depend as heavily as DNA viruses on the cell cycle.
Group IV: Single-stranded, Positive-sense, RNA viruses
All RNA viruses defined as positive-sense can be directly accessed by the host ribosomes immediately to form proteins. Examples of this class include the families Astroviridae, Caliciviridae, Coronaviridae, Flaviviridae, Picornaviridae, Arteriviridae, and Togaviridae. These can be divided into two groups, both of which reproduce in the cytoplasm.
- Viruses with polycistronic mRNA: In this group, the genome RNA forms the mRNA which is translated into a polyprotein product that is converted to the mature proteins. This means that the gene can use a few methods in which to produce proteins from the same strand of RNA, all in the sake of reducing the size of its gene.
- Viruses with complex transcription, for which subgenomic mRNAs, ribosomal frameshifting, and proteolytic processing of polyproteins may be used. All of which are different mechanisms with which virus can produce proteins from the same strand of RNA.
Group V: Single-stranded RNA viruses – Negative-sense
All the genes defined as negative-sense cannot be directly accessed by host ribosomes to immediately form proteins. Instead, they must be transcribed by viral polymerases into a “readable” form, which is the positive-sense reciprocal. Examples in this class include the families Arenaviridae, Orthomyxoviridae, Paramyxoviridae, Filoviridae, and Rhabdoviridae (the latter of which includes the rabies virus). These can also be divided into two groups:
- Viruses containing nonsegmented genomes: In these, the first step in replication is transcription from the negative stranded genome by the viral RNA-dependent RNA polymerase to form monocistronic mRNAs that code for the various viral proteins. A positive-sense genome copy is then produced that serves as a template for the production of the negative strand genome. Replication occurs within the cytoplasm.
- Viruses with segmented genomes: Here, replication occurs in the nucleus and the viral RNA-dependent RNA polymerase forms monocistronic mRNAs from each segment of the genome. The largest difference between the two is the occurance of replication in different locations.

Group VI: Positive-sense single-stranded RNA viruses that replicate through a DNA intermediate
- A well-studied family of this class of viruses includes the retroviruses.
- One defining feature of this group is the use of reverse transcriptase to convert the +sense RNA into DNA.
- They do not use RNA as templates to make proteins, but use DNA to create the templates, which is spliced into the host genome using integrase. Replication then commences with the help of the host cell’s polymerases.
- Examples: Retroviruses
Group VII: Double-stranded DNA viruses that replicate through a single-stranded RNA intermediate
- This group of viruses has a double-stranded, gapped genome that is subsequently filled in to form a covalently closed circle (cccDNA) that serves as a template for production of viral mRNAs and a subgenomic RNA.
- The pregenome RNA serves as template for the viral reverse transcript to produce the DNA genome.
- Example: Hepatitis B virus (which is in the Hepadnaviridae family)
ICTV virus classification
The International Committee on Virus Taxonomy founded in 1966, designs and implements the rules for naming and classifying viruses. ICTV works under the umbrella of the International Union of Microbiological Societies with the task of developing, refining, and maintaining a universal virus taxonomy.
ICTV’s definition of virus species
In July 2013 ICTV declared virus species in such a way that “A species is a monophyletic group of viruses whose properties can be distinguished from other species by multiple criteria.” These are physical particles composed of DNA and proteins, produced by genetic or biological evolution. Concepts of species and taxa in viral classification are abstracts taken by the rational system of classification for ease to understand.
Since 1971, ICTV has reported eight reports about virus classification which are summarized below.
| Report | Reference | Reporting ICTV Proceedings at the International Congress of Virology held in: | Content |
| First | Wildy (1971) | Helsinki, 1968 | Total 43 families and groups |
| Second | Fenner (1976) | Budapest, 1971 and Madrid, 1975 | Total 47 families and groups |
| Third | Matthews (1979) | The Hague, 1978 | Total 50 families and groups |
| Fourth | Matthews (1982) | Strasbourg, 1981 | Total 54 families and groups |
| Fifth | Francki et al. (1991) | Sendai, 1984, Edmonton, 1987, and Berlin, 1990 | Total 2420 viruses belonging to 73 families or groups |
| Sixth | Murphy et al. (1995) | Glasgow, 1993 | Total 3,600 virus species belonging to 1 order, 50 families, 9 subfamilies, 164 genera. |
| Seventh | van Regenmortel et al. (2000) | Jerusalem, 1996 | Total 1550 species belonging to 3 orders, 63 families, 9 subfamilies, 240 genera. |
| Eighth | Fauquet et al. (2005) | Sydney, 1999 and Paris, 2002 | Total 1898 species belonging to 3 orders, 73 families, 11 subfamilies, 289 genera. |
Related posts:
- Definition, structure and functions of viruses
- General characteristics of viruses
- Replication in viruses
- Examples of good viruses in health
- Benefits of viruses
